New Vaccine Advances Could Help Against More Viral Illnesses
Some of the same researchers at The University of Texas at Austin who created a key to all coronavirus vaccines used in the U.S. have made a similar advance against the human metapneumovirus (hMPV), one of a handful of remaining respiratory viruses for which there is currently no vaccine.

Ching-Lin Hsieh and Jason McLellan are among the UT Austin scientists who have engineered a protein of the human metapneumovirus for use in vaccines. Credit: Vivian Abagiu
The work is described in a paper published today in Nature Communications.
For elderly people, the immunocompromised and babies, COVID-19 isn't the only dangerous respiratory illness. Vaccines are still needed for many viruses that cause hospitalizations and thousands of deaths in vulnerable populations each year.
Structure-based vaccines are relatively new and, like mRNA technology, have proved to be game changing for the fight against infectious diseases, including COVID-19. Using highly detailed images of viral proteins and other parts of a virus, scientists learn to develop the ideal blueprint to include in a vaccine and train the body to recognize and fight off invading pathogens.
In the new paper, professor of molecular biosciences Jason McLellan and his team describe how to engineer the hMPV fusion protein to increase its stability and usefulness in vaccines, specifically by locking it in what scientists call its prefusion conformation, or the shape of the protein before the virus enters a host cell. Without the modifications, the protein shape-shifts and assumes an alternative conformation that elicits a sixfold-lower immune response when injected into mice. The scientists stripped the viral protein of its shape-shifting ability by introducing some genetic modifications that act like molecular staples and lock the structure in the optimal form for use in a vaccine.
"Vaccines being developed with structural information and guided by that structural information really represent a wave of the future," McLellan said. "Structural biology, protein design and engineering are used not just to fight viruses, but also other pathogens, like bacteria, and companies are ramping up their investments in these areas."
The paper's first author is postdoctoral scientist Ching-Lin Hsieh. Other authors are graduate students Scott Rush, Chia-Wei Chou and Whitney Pickens of UT Austin and scientists Vicente Mas and Concepcion Palomo of Instituto de Salud Carlos III in Spain. Hsieh, Rush and McLellan have filed a patent application on the protein technology described in the paper.
The company Icosavax (Nasdaq: ICVX) describes plans on its website to begin clinical trials later this year for a vaccine that uses the protein mutations McLellan and the Texas team worked on for hMPV, as well as technology McLellan and colleagues at the National Institutes of Health (NIH) developed against the respiratory syncytial virus (RSV) several years ago to guard against both diseases.
McLellan is a pioneer of using structural biology and protein engineering for vaccine development. With colleagues at the NIH, he and his team designed the version of the spike protein found in many vaccines against COVID-19, and that design also uses stabilizing mutations. In 2013, he and colleagues determined how to modify a protein of RSV for use in vaccines, and vaccines based on that work are now entering the final stage of clinical trials. If approved, the vaccines would be the first against RSV.
"This is why I chose to go into science, to try to make an impact on human health," McLellan said. What initially seemed like a long shot for his research has resulted instead in highly effective vaccines. At the one-year mark, COVID-19 vaccines have been estimated to have saved 1.1 million lives in the United States alone, while preventing 10.3 million U.S. hospitalizations.
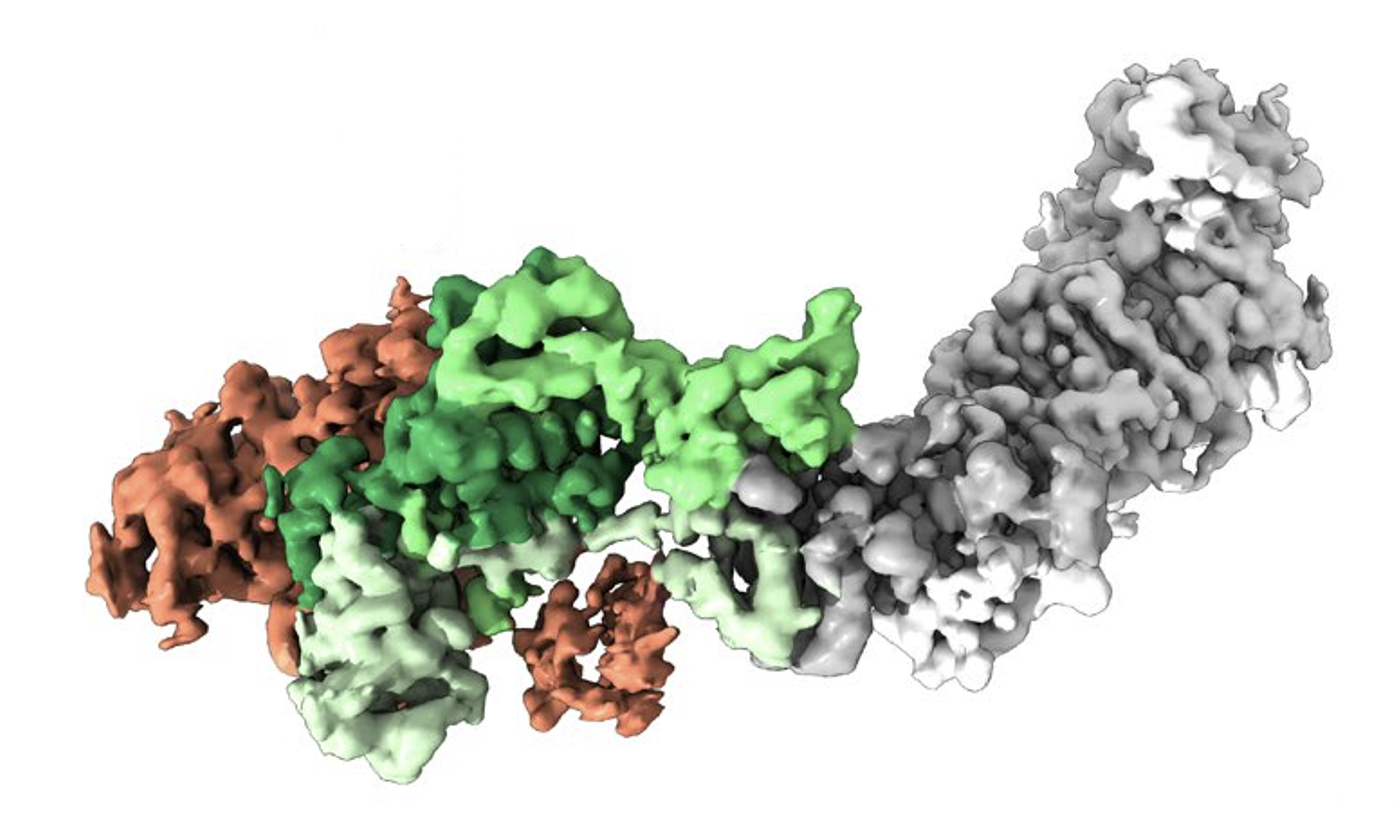
The structure of the human cytomegalovirus, a virus for which there is currently no vaccine. Credit: McLellan Lab
McLellan and his team also published research Friday in the journal Science Advances, identifying for the first time the structure of a key infectious part of the human cytomegalovirus (HCMV), another virus for which there is no vaccine and that can cause severe illness in people with impaired immune systems and in developing fetuses. With colleagues at The University of Texas Health Science Center at Houston, McLellan's team uncovered part of the process by which HCMV infects host cells and how naturally produced antibodies neutralize the virus— insights that could help scientists develop new vaccines or therapeutics.
Using a state-of-the-art imaging technique called cryo-EM, the researchers for the first time created a 3D atomic-scale map, or structure, of a protein complex called the HCMV Pentamer that helps the HCMV infect host cells by binding to a receptor called NRP2. They also created structures of the protein complex bound to four different neutralizing antibodies. The imaging data were collected at the Sauer Structural Biology Laboratory at UT Austin.
"As a team, we synergized the strengths of UT Austin's structural biology and UTHealth Houston's antibody discovery, and it has led to the success of this project," said Zhiqiang An, a professor, Robert A. Welch Distinguished University Chair in Chemistry, and director of Texas Therapeutics Institute at UTHealth Houston. "Such cross-institutional collaborations within the UT System should be encouraged and expanded."
The Science Advances study's co-first authors are Daniel Wrapp, a former graduate student in McLellan's lab and now a postdoctoral fellow at the Duke Human Vaccine Institute, and Xiaohua Ye, a former postdoctoral researcher and now instructor at UTHealth Houston.
The two studies were funded by The Welch Foundation. Research was also supported, in part, by grants from Merck & Co. Inc. and the Texas Emerging Technology Fund. The Sauer Structural Biology Laboratory is supported by Tito's Handmade Vodka, the UT Austin College of Natural Sciences and the Cancer Prevention and Research Institute of Texas (CPRIT). Argonne at the University of Chicago, which was involved in the work published in Nature Communications, is part of the U.S. Department of Energy. McLellan holds the Robert A. Welch Chair in Chemistry at UT Austin and is a corresponding author for both new studies.
The University of Texas at Austin is committed to transparency and disclosure of all potential conflicts of interest. The authors submitted required financial disclosure forms with the university and hold intellectual property rights that may yield revenue from the discovery described in this research.