COVID-19 Vaccines with UT Ties Arrived Quickly After Years in the Making
When the first COVID-19 vaccine trial in the U.S. began on March 16, history was being made. Never before had a potential vaccine been developed and produced for human trials so quickly—just 66 days since scientists published the genome sequence of the virus that causes the disease. This blindingly fast effort was only possible because a group of scientists and their partners in industry had already invested years in laying the groundwork.
Locked and Loaded
In 2008, current UT Austin associate professor of molecular biosciences Jason McLellan was just beginning a postdoctoral fellowship in the National Institutes of Health's Vaccine Research Center (VRC). Working with the lab of Dr. Barney Graham, he was partnering on the search for a vaccine to combat a respiratory virus called RSV. Lessons Graham and McLellan learned in the process not only led to a potential vaccine for that virus; it also provided essential clues for finding a vaccine for the current novel coronavirus.
In both viruses, a key protein responsible for infecting cells changes shape before and after infection. If the immune system encounters the protein in the first shape, it makes potent antibodies, but not so if the protein has taken on the second shape.
When they worked together at the VRC, McLellan and Graham used cutting-edge technology to determine the structure of RSV's infection-causing protein at the atomic-level, then they re-engineered the protein to take away its shape-shifting ability. Locking it in the shape that elicits the best antibodies led to a new type of vaccine candidate, designed especially to address the structure- shifting problem in RSV and leading to a vaccine currently in Phase 2 human trials.
The same structure-locking strategy would prove useful in the fight against the virus that causes COVID-19.
"Our first time testing these stabilized molecules in animals, the response was tenfold higher than anything anyone had ever seen before," McLellan said. "And at that point, we're thinking, 'This is it. We've got it.' That was exciting."
After outbreaks of two different coronaviruses early in the century, MERS-CoV and SARS-CoV, Graham and McLellan began researching ways to combat coronaviruses, too. Like RSV, coronaviruses have a key protein on the surface that changes shape before and after infecting a cell. In this case, the key is the spike protein; like the studs on a punk rocker's dog collar, these crown-like spikes inspired the name coronavirus.
"We felt that we could apply the lessons of structure-based vaccine design to coronaviruses, too," McLellan said.
He left to start his own lab as a faculty member, first at Dartmouth College in 2013 then at The University of Texas at Austin in 2018, when he was recruited in part through UT System's STARS program. McLellan and his team of collaborators were receiving attention for having discovered ways to lock different coronavirus spike proteins into their pre-fusion shape.
Early on, McLellan hired a postdoctoral researcher, Nianshuang Wang, who in graduate school had studied MERS. Wang identified genetic mutations that would stabilize the shape-shifting spike protein for that coronavirus – and, as it turned out, other coronaviruses, too. The key was making small genetic modifications to the gene sequence that encodes for the protein, something they filed a patent for in 2017. As Brooke Jarvis summarized in an article in WIRED magazine, the team learned "they could use carefully engineered genetic mutations as if they were staples, locking down regions of the spike that wanted to move around by binding them to regions that did not."
This trick that had worked for RSV, SARS-CoV and MERS-CoV proteins would become a key element of a vaccine candidate for the next coronavirus to emerge: SARS-CoV-2.
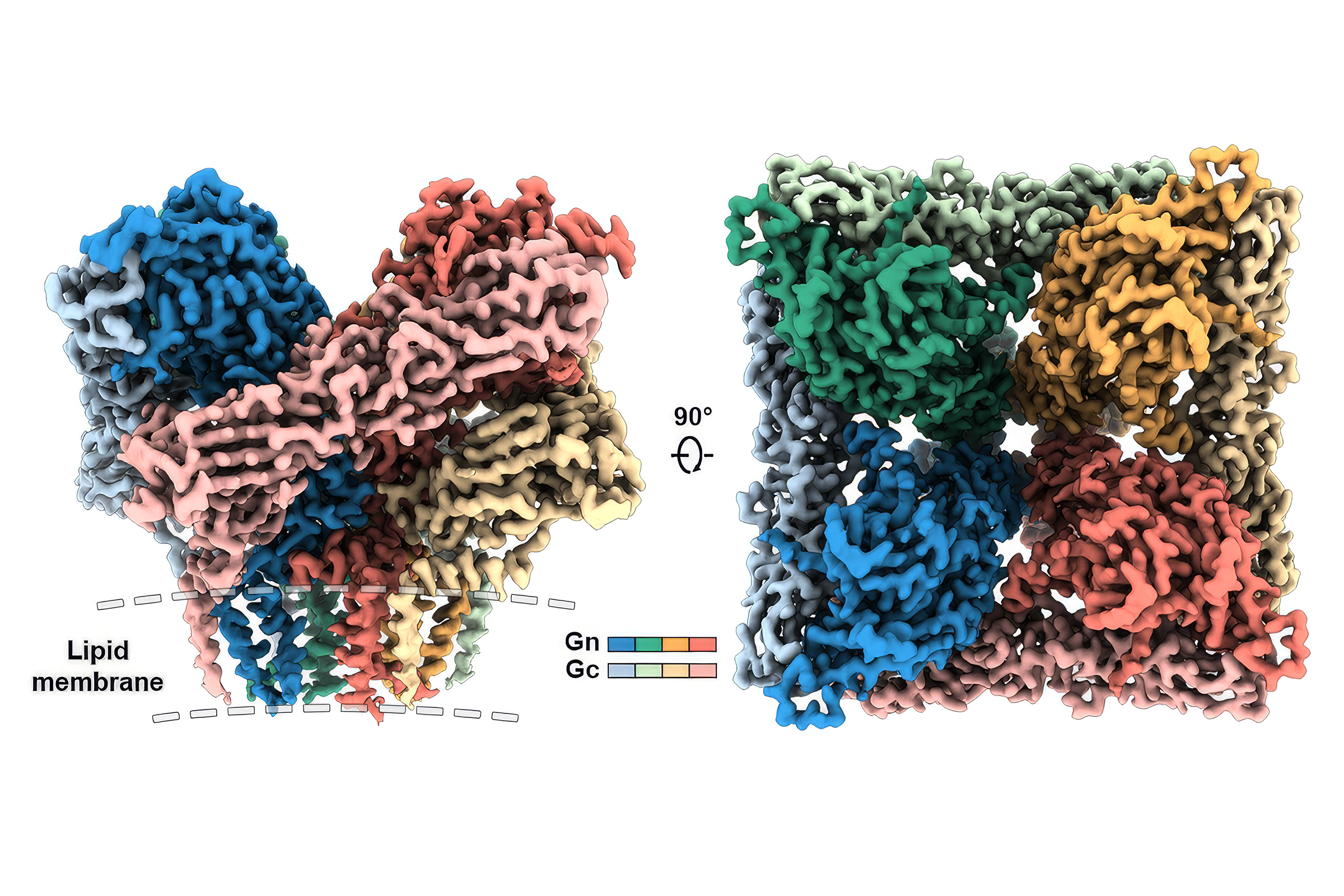
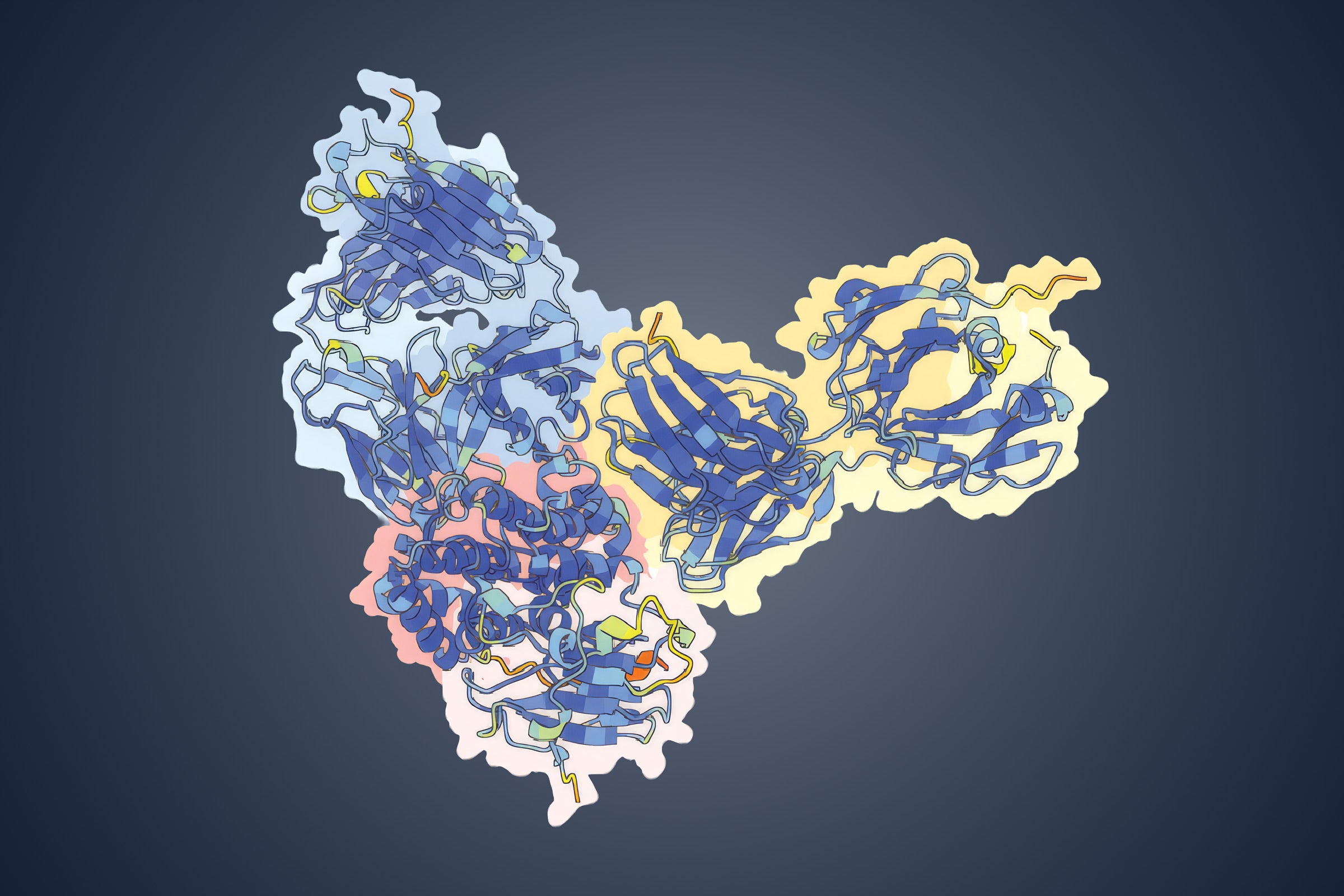
This is a 3D atomic scale map, or molecular structure, of the 2019-nCoV spike protein. The protein takes on two different shapes, called conformations—one before it infects a host cell, and another during infection. This structure represents the protein before it infects a cell, called the prefusion conformation. It also has some small stabilizing mutations that lock it into this shape. Credit: Jason McLellan/University of Texas at Austin.
The Race is On
In January 2020, McLellan was snowboarding with his family in Utah when Barney Graham, now deputy director of the VRC, called to tell him the disease spreading in Wuhan, China was apparently a coronavirus.
"Are you ready to get to work on it?" Graham asked, according to Laura Howes at Chemical & Engineering News. McLellan immediately sent a message to his team: "We're going to race as soon as we get the sequence."
They didn't have to wait long. On January 10, researchers in China published a draft genome sequence of the new virus.
"As soon as we knew this was a coronavirus, we felt we had to jump at it to get this structure," McLellan said, "We knew exactly what mutations to put into this, because we've already shown these mutations work for a bunch of other coronaviruses."
Thanks to their experience with MERS-CoV and SARS-CoV, McLellan and his team, which includes Wang and graduate student Daniel Wrapp, had three advantages that allowed them to blast through the early steps of vaccine development.
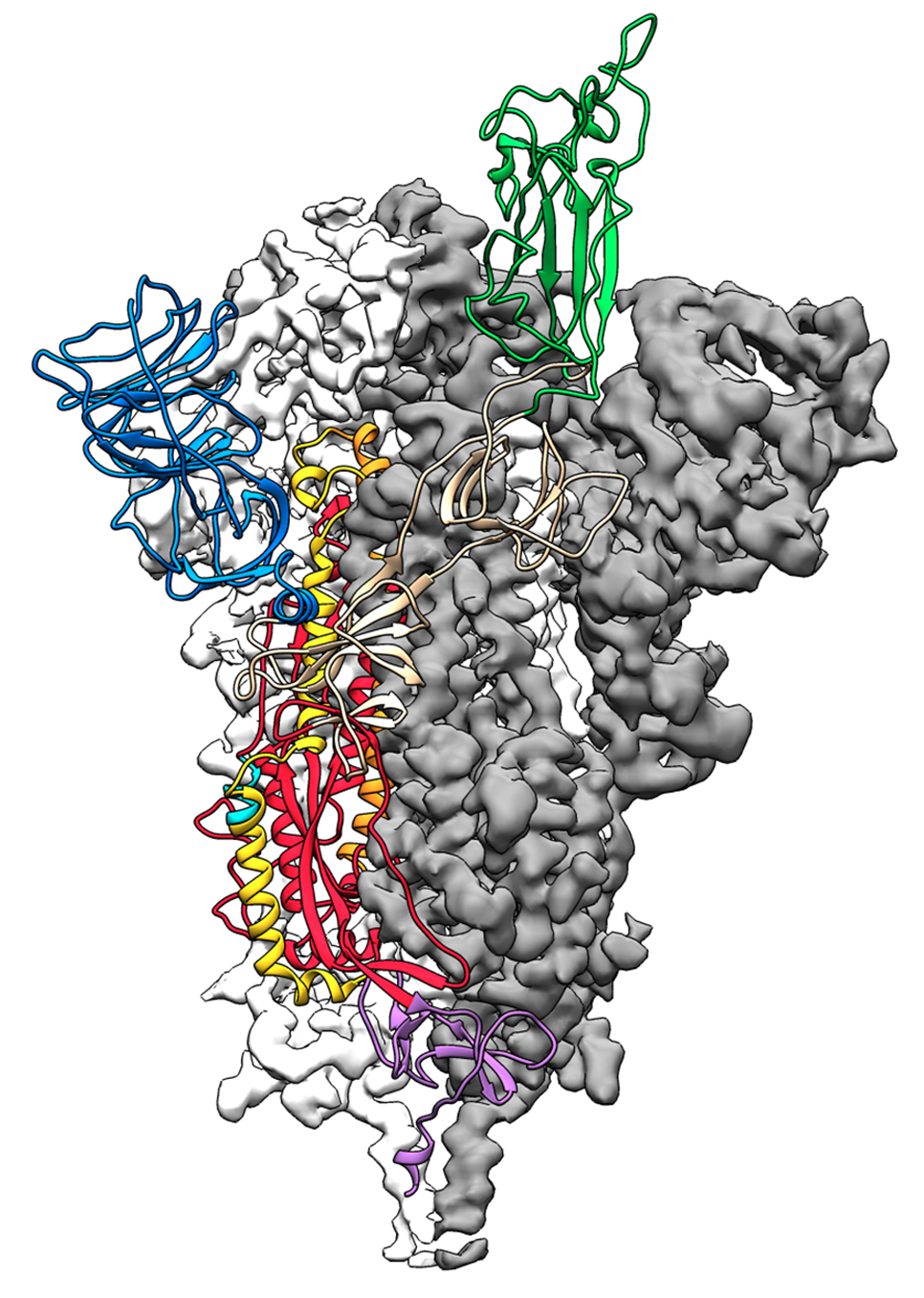
First, the team's past work in stabilizing the spike proteins from other coronaviruses gave them a toolkit of modifications that had a high chance of working for the new virus. Instead of a painful process of trial and error, the researchers could confidently design necessary mutations rapidly (in about a day). Identifying the section of the genome that codes for the spike protein and creating mutations that would lock it into the pre-fusion shape helped the team to do two things before any other researchers in the world could: figure out the three-dimensional structure of the protein and begin to develop a safe version of it for the body to recognize and respond to as part of a dangerous virus. In other words, the researchers could kill two birds with one stone—take a detailed portrait of the protein for research and simultaneously create a possible vaccine antigen to protect people.
From left: Jason S. McLellan, associate professor of molecular biosciences, Daniel Wrapp, graduate student, and Nianshuang Wang, research associate, pose for a photo in the McLellan Lab at The University of Texas at Austin Monday Feb. 17, 2020. Credit: Vivian Abagiu.
Second, McLellan's lab members and their collaborators at the NIH had already learned how to take the genetic code for a modified spike protein, insert it into human cells in a culture and coax the cells to produce pure samples of the protein. That very first weekend, Wang was able to turn around the design for a gene construct necessary to map the spike protein, and by the end of January, Wrapp had harvested and purified the spike protein, helping to pave a path to the creation of a synthetic version of the protein.
Third, the UT researchers had immediate access to—and several years of practice using—state-of-the-art imaging technology known as cryogenic electron microscopy (cryo-EM). They were among the first users of UT Austin's new Sauer Laboratory for Structural Biology, which houses cryo-EM technology capable of making atomic-scale 3D models of cellular structures, molecules and viruses.
Partly funded through the state's unique Cancer Prevention and Research Institute of Texas, the Sauer Lab is equipped with one of the best electron microscopes in the world. The researchers were able to reconstruct the molecular structure of their stabilized spike protein in about a week and soon after published the structure of the SARS-CoV-2 spike protein in the journal Science.
"We ended up being the first ones in part due to the infrastructure at the Sauer Lab," McLellan said. "It highlights the importance of funding basic research facilities."
Vaccine makers the world over contacted the lab, as this map was a big milestone, providing crucial information for dozens of efforts. The process of mapping the 3D structure of the protein required stabilizing it, too. That made their version of the protein in essence a vaccine antigen, something you could potentially give to patients to protect them from the virus.
The UT Austin team along with the team from NIH, which included Graham and Kizzmekia Corbett, scientific lead for the coronavirus team at the VRC, filed a joint patent application on their stabilized spike protein soon after.
Nianshuang Wang, research associate, at work in the lab Monday Feb. 17, 2020 at The University of Texas at Austin. Credit: Vivian Abagiu.
Plug and Play
Corbett and Graham at the VRC verified that the stabilized version of the spike protein generated potent antibodies in mice. Using a "plug and play" approach, the Boston-based biotech firm Moderna incorporated the genetic sequence for this version of the protein in a new type of vaccine platform it had developed. This vaccine didn't involve injecting a person with a weakened or dead virus, or even a part of a virus, like a traditional vaccine but instead injecting messenger RNA—bits of genetic code that tell cells to produce specific proteins and how to make them. Patients receiving these injections would enlist their own cells to become factories, producing not human proteins, but viral proteins that would train the body's immune system to recognize dangerous viruses without ever actually encountering them.
Graham had previously worked with Moderna on a potential MERS vaccine. Pairing the scientific teams' spike protein with the company's mRNA vaccine platform led to the creation of a vaccine called mRNA-1273. The first batch of the vaccine was completed on February 7. Then on March 2, the US Food and Drug Administration approved a Phase 1 study in humans.
Two weeks later, the first of the 45 adult volunteers were injected with the new vaccine, marking the first vaccine trials in the U.S. aimed at halting a terrifying pandemic that has already killed more than half a million people globally.
"It was a lot faster than even the fastest one we'd previously done," said Barney Graham, according to Nature. Because of the earlier research on SARS and MERS, coronaviruses were probably the only viral family for which that was possible, he added.
The vaccine candidate mRNA-1273 has shown promising results in a phase 1 human clinical trial and is currently in Phase 2 trials. While there are currently about 23 vaccine candidates in human trials around the world, Moderna's vaccine is the farthest along in the U.S., with a Phase 3 trial in 30,000 subjects set to begin in late July. Experts have said the world will know whether the Moderna vaccine is a success as early as this fall.
More in the Pipeline
Moderna's is not the only vaccine under development and in human trials that McLellan and his researchers have had a hand in. A subunit vaccine developed by Novavax consists of purified spike proteins stabilized in the prefusion conformation using the same mutations; Johnson and Johnson's vaccine and the vaccine developed by Pfizer/BioNTech all also use the team's stabilized spike protein.
Meanwhile, the team in Texas has kept plugging away, creating an improved version of the protein in more recent weeks. The Bill & Melinda Gates Foundation has contributed to the development of the technology through a grant in the interest of making vaccines accessible to people in lower-income countries. Vaccine companies with different platform technologies will have the ability to test and further develop COVID vaccines that use this improved version of the spike protein.
If these vaccines are successful, a small team of UT Austin researchers will have played a critical role in solving arguably the world's most urgent scientific challenge.
The University of Texas at Austin is committed to transparency and disclosure of all potential conflicts of interest. The authors submitted required financial disclosure forms with the university and hold intellectual property rights that may yield revenue from discoveries described in this research. This article was updated in January 2021, as vaccines using the Texas team's stabilized spike protein came into circulation.

Nianshuang Wang, research associate, right, and Daniel Wrapp, graduate student, left, review cryo-EM images in the The Sauer Structural Biology Laboratory Monday Feb. 17, 2020 at The University of Texas at Austin. Credit: Vivian Abagiu.